pacemaker workflow#
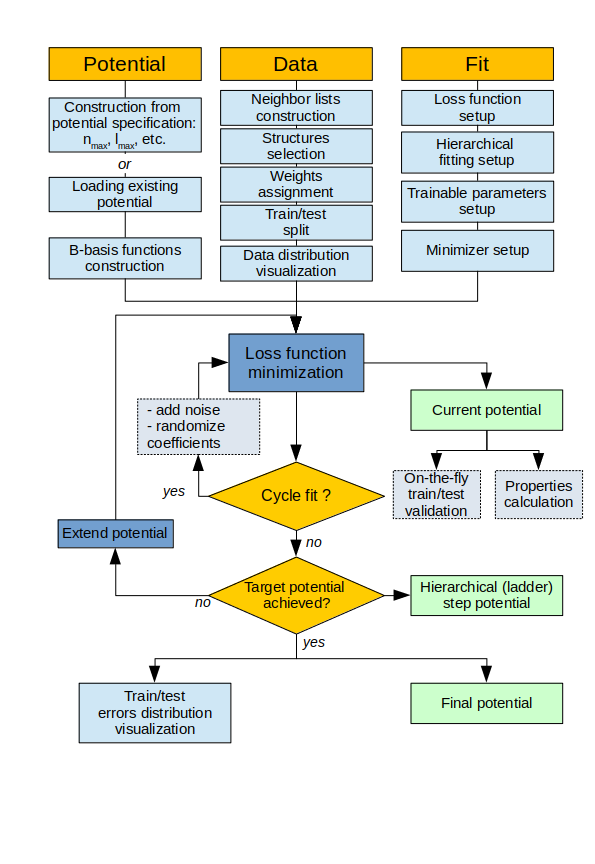
The pacemaker workflow is described in the following and summarized in the figure above.
pacemakerstarts by constructing the potential according to the user specified basis configuration (\(\nu\) -order, \(n_\textrm{max}\), \(l_\textrm{max}\), etc.) or loads it from an available potential file. Then the B-basis functions are constructed, to this end generalized Clebsch-Gordan coefficients are set up for generating product basis functions that are invariant with respect to rotation and inversion.- Then
pacemakerconstructs the neighborlist for all structures in the dataframe. The neighborlist can be added to the reference dataframe for a fast restart of future parameterization runs. - Next the weights for each structure and atom as required by the loss function are set up.
pacemakerprovides different weighting schemes. The weights are then added to the reference dataframe. Weights may also be added directly to the reference dataframe, so that the user has full control over the weights for each structure and force. pacemakersplits the dataset for training and for testing.- The further specification of \(\mathrm{L}_1\), \(\mathrm{L}_2\) and radial smoothness \(w_0, w_1, w_2\) regularization contributions and the relative weight \(\kappa\) of energy and force errors enables
pacemakerto set up the loss function. - The hierarchical basis extension is setup as ladder fitting scheme if requested by the user.
- The optimization of the loss function can be carried out with different optimizers and optimization strategies. For each optimization step
pacemakerstores the current potential and computes error metrics for energies and forces. In addition, external Python code can be called to perform specific calculations for advanced on-the-fly validation. - If requested, optimization is repeated with intermediate randomization of the training parameter.
- During and at the end of loss function optimization
pacemakerprovides outputs for assessing the quality and convergence of a parameterization. pacemakerstores (and loads) the ACE potentials in a transparent YAML file format.